Research in my laboratory is focused on understanding the molecular mechanisms, biological functions, and evolutionary conservation of various coronavirus-host interactions. Relying on a combination of cutting-edge cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) technique with complementary biophysical, biochemical, cellular, genomic, and computational approaches, we are developing new conceptual frameworks and design novel platforms and tools to characterize and identify new host and viral factors involved in the novel mechanisms of gene regulation.
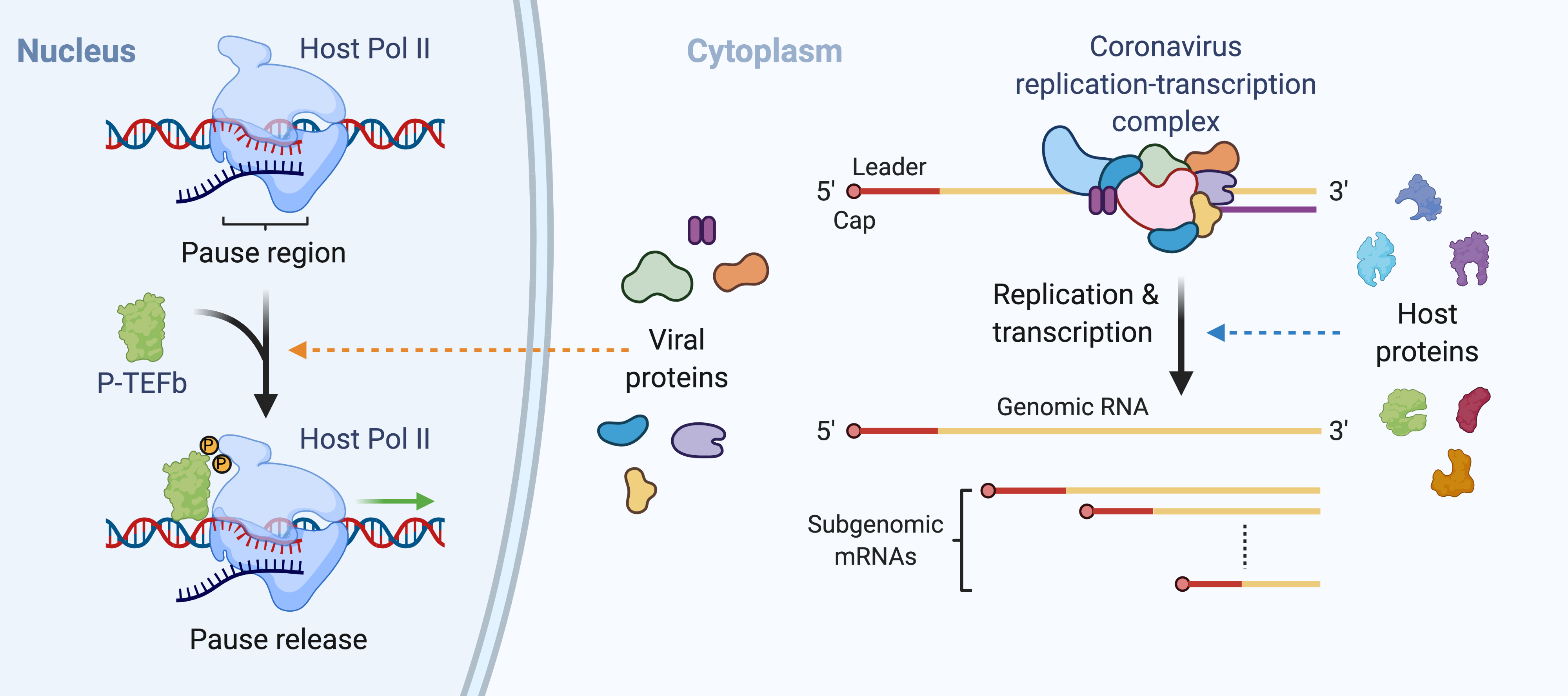
Modulation of host transcriptional landscape by SARS-CoV-2 proteins. Host gene expression is intricately regulated at the stage of promoter-proximal pausing of RNA polymerase II (Pol II), with elongation reactivated by the positive transcription elongation factor b (P-TEFb). Very recent studies indicate that some SARS-CoV-2 proteins interact with critical host factors that control the accessibility of P-TEFb to Pol II, raising the intriguing questions: what are the functions of these interactions and what mechanisms govern these functions? To understand how the SARS-CoV-2 proteins interact with host transcriptional regulators and how these virus-host interactions may perturb the reactivation of paused Pol II, thus inhibiting the expression of host genes, especially those involved in antiviral responses, we are developing novel ribonucleoprotein (RNP) complex reconstitution strategies and employing cutting-edge cryo-EM/cryo-ET reconstruction pipelines and use them to determine how and in what contexts the viral proteins interact with the critical P-TEFb regulating factors to regulate Pol II reactivation. In addition, we are examining the cellular impacts of these SARS-CoV-2 protein on host transcriptional landscapes in the context of SARS-CoV-2 infection.
Regulation of SARS-CoV-2 replication and transcription by host proteins. Coronavirus replication and transcription are carried out by the multi-subunit viral replication-transcription complex (RTC). An array of host proteins that are critically involved in antiviral defense were found to interact with different subunits of the SARS-CoV-2 RTC and affect viral replication. Intriguingly, some of these host-virus interactions were shown to suppress SARS-CoV-2 in previously unknown manners, suggesting new approaches for antiviral intervention. To elucidate how host proteins sequester the viral replication-transcription machine to restrict viral gene expression, we are determining the molecular mechanisms and evolutionary conservation of these host-virus interactions and using a novel SARS-CoV-2 reporter replicon system to examine their consequences for viral gene expression. In addition, we are also developing an innovative capture-identification-visualization platform discover new host factors that mediate novel viral replication/transcription-inhibiting mechanisms.
